Adjacency graph of classes in packages
adjacencyOfClasses.RdGet inheritance graph of classes in one or more packages.
Usage
adjacencyOfClasses(packages, externalSubclasses = FALSE,
result = c("default", "matrixOfPairs", "adjacencyMatrix"),
Wolfram = FALSE)Arguments
- packages
names of one or more packages, a character vector
- externalSubclasses
if
TRUE, exclude subtrees of classes not defined in any of the packages listed in argumentpackages.- result
format of the result, can be missing or one of
"default","matrixOfPairs","adjacencyMatrix", see Details.- Wolfram
if
TRUE, print a suitable graph expression to be run by Mathematica, see Details.
Details
adjacencyOfClasses computes a graph representation of the
dependencies of S4 classes defined in one or more packages (as
specified by argument package) and returns a list. The
contents of the list returned by adjacencyOfClasses depend on
argument result. Partial matching is used for the value of
argument result, e.g., "adj" is equivalent to
"adjacencyMatrix".
If externalSubclasses = FALSE, the default, subclasses defined
outside the requested packages are excluded. This is typically what
the user will be looking for. To get a complete tree, set
externalSubclasses to TRUE.
The S4 classes are represented by the vertices of the graph.
Component "vertices" of the result gives them as a character
vector. References below to the \(i\)th class or vertex
correspond to the order in this vector. No attempt is made to arrange
the vertices in a particular order. An empty list is returned if this
vector is empty.
If result is missing or "default", the edges of the
graph are represented by a character vector. Each edge is represented
by a string with an arrow "->" from a superclass to a
subclass. Here is an example that shows that this package defines one
class, which is a subclass of "list":
adjacencyOfClasses("gbutils")
##: $vertices
##: [1] "objectPad" "list" ##: $edges
##: [1] "list -> objectPad"This illustrates the effect of argument "externalSubclasses":
adjacencyOfClasses("gbutils", externalSubclasses = TRUE)
##: $vertices
##: [1] "objectPad" "list" "vector" ##: $edges
##: [1] "list -> objectPad" "vector -> list" The edge, "vector -> list" was omitted in the previous example since
this relationship is defined elsewhere. This resulted in class
"vector" being dropped also from the vertices, since it is not
defined in "gbutils" and none of the remaining edges contains
it.
If result is "matrixOfPairs", the edges of the
graph are represented by a character matrix with two columns, where each row
represents an edge from the element in the first column to the element
in the second. In this example there is one edge, so the matrix
contains one row:
adjacencyOfClasses("gbutils", result = "matrixOfPairs")
##: $vertices
##: [1] "objectPad" "list" ##: $edges
##: [,1] [,2]
##: [1,] "list" "objectPad"If result is "adjacencyMatrix", the adjacency matrix of
the graph is in component "AM" of the returned list. Element \((i,j)\) of
this matrix is equal to one, if the \(j\)th class is a superclass
to the \(i\)th. In other words, the \(j\)th column gives the
superclasses of the \(i\)th class. Here the element in position
(1,2) is non-zero, so "list" is the superclass of "objectPad":
adjacencyOfClasses("gbutils", result = "adjacencyMatrix")
##: $vertices
##: [1] "objectPad" "list" ##: $AM
##: objectPad list
##: objectPad 0 1
##: list 0 0Note that including the vertices in the result is not redundant, since some may not be in any edge. This can happen if a class does not have any superclasses and subclasses.
As described above the result is not converted to a graph object but it can be fed to functions provided by a number of R packages.
An additional option is to use argument Wolfram. If
Wolfram is TRUE, a suitable Mathematica command is
printed. It can be evaluated in a Mathematica session (e.g., by
copy/paste) to produce a graphical representation of the graph and/or
be manipulated further by it. This feature is a side effect, the
return value of adjacencyOfClasses is as controlled by the
other arguments. For example, the return value below is as without
argument "Wolfram" but, in addition, the printed line defines a
Wolfram language graph in terms of its vertices and edges:
adjacencyOfClasses("gbutils", Wolfram = TRUE)
##: Graph[{objectPad,list}, {list -> objectPad}, VertexLabels -> Automatic] ##: $vertices
##: [1] "objectPad" "list" ##: $edges
##: [1] "list -> objectPad"Setting result = "adjacencyMatrix" in the last R command
exports the graph in terms of its adjacency matrix:
adjacencyOfClasses("gbutils", Wolfram = TRUE, result = "adjacencyMatrix")
##: AdjacencyGraph[{objectPad,list}, {{0, 0},
##: {1, 0} }
##: , VertexLabels -> Automatic] ##: $vertices
##: [1] "objectPad" "list" ##: $AM
##: objectPad list
##: objectPad 0 1
##: list 0 0Value
a list with some of the following components (as described in Details):
- vertices
a character vector of S4 class names,
- edges
the edges of the graph, in the format controlled by argument
results(not present whenresultis equal to"adjacencyMatrix"),- AM
the adjacency matrix of the graph (present only when
resultis"adjacencyMatrix").
References
Gentleman R, Whalen E, Huber W, Falcon S (2017). graph: A package to handle graph data structures. R package version 1.56.0.
Hansen KD, Gentry J, Long L, Gentleman R, Falcon S, Hahne F, Sarkar D (2017). Rgraphviz: Provides plotting capabilities for R graph objects. R package version 2.22.0.
Maechler M (2015). classGraph: Construct Graphs of S4 Class Hierarchies. (partly based on code from Robert Gentleman) R package version 0.7-5, https://CRAN.R-project.org/package=classGraph.
See also
?methods::classesToAM which is used for the main computation
here,
Maechler (2015) for a suite of related functions. Gentleman et al. (2017) for creation and manipulation of graphs, and Hansen et al. (2017) for visualisation of graphs.
Examples
adjacencyOfClasses("gbutils")
#> $vertices
#> [1] "objectPad" "list"
#>
#> $edges
#> [1] "list -> objectPad"
#>
adjacencyOfClasses("gbutils", TRUE)
#> $vertices
#> [1] "objectPad" "list" "vector"
#>
#> $edges
#> [1] "list -> objectPad" "vector -> list"
#>
adjacencyOfClasses("gbutils", FALSE, "matrixOfPairs")
#> $vertices
#> [1] "objectPad" "list"
#>
#> $edges
#> [,1] [,2]
#> [1,] "list" "objectPad"
#>
adjacencyOfClasses("gbutils", TRUE, "matrixOfPairs")
#> $vertices
#> [1] "objectPad" "list" "vector"
#>
#> $edges
#> [,1] [,2]
#> [1,] "list" "objectPad"
#> [2,] "vector" "list"
#>
adjacencyOfClasses("gbutils", FALSE, "adjacencyMatrix")
#> $vertices
#> [1] "objectPad" "list"
#>
#> $AM
#> objectPad list
#> objectPad 0 1
#> list 0 0
#>
adjacencyOfClasses("gbutils", TRUE, "adjacencyMatrix")
#> $vertices
#> [1] "objectPad" "list" "vector"
#>
#> $AM
#> objectPad list vector
#> objectPad 0 1 0
#> list 0 0 1
#> vector 0 0 0
#>
## as above, also represent the graph using the edges
adjacencyOfClasses("gbutils", Wolfram = TRUE)
#> Graph[{objectPad,list}, {list -> objectPad}, VertexLabels -> Automatic]
#>
#> $vertices
#> [1] "objectPad" "list"
#>
#> $edges
#> [1] "list -> objectPad"
#>
adjacencyOfClasses("gbutils", TRUE, Wolfram = TRUE)
#> Graph[{objectPad,list,vector}, {list -> objectPad, vector -> list}, VertexLabels -> Automatic]
#>
#> $vertices
#> [1] "objectPad" "list" "vector"
#>
#> $edges
#> [1] "list -> objectPad" "vector -> list"
#>
## here the graph is represented by the adjacency matrix:
adjacencyOfClasses("gbutils", FALSE, "adjacencyMatrix", Wolfram = TRUE)
#> AdjacencyGraph[{objectPad,list}, {{0, 0},
#> {1, 0} }
#> , VertexLabels -> Automatic]
#>
#> $vertices
#> [1] "objectPad" "list"
#>
#> $AM
#> objectPad list
#> objectPad 0 1
#> list 0 0
#>
adjacencyOfClasses("gbutils", TRUE, "adjacencyMatrix", Wolfram = TRUE)
#> AdjacencyGraph[{objectPad,list,vector}, {{0, 0, 0},
#> {1, 0, 0},
#> {0, 1, 0} }
#> , VertexLabels -> Automatic]
#>
#> $vertices
#> [1] "objectPad" "list" "vector"
#>
#> $AM
#> objectPad list vector
#> objectPad 0 1 0
#> list 0 0 1
#> vector 0 0 0
#>
if(requireNamespace("graph", quietly = TRUE) &&
requireNamespace("Rgraphviz", quietly = TRUE)) withAutoprint({
## another package
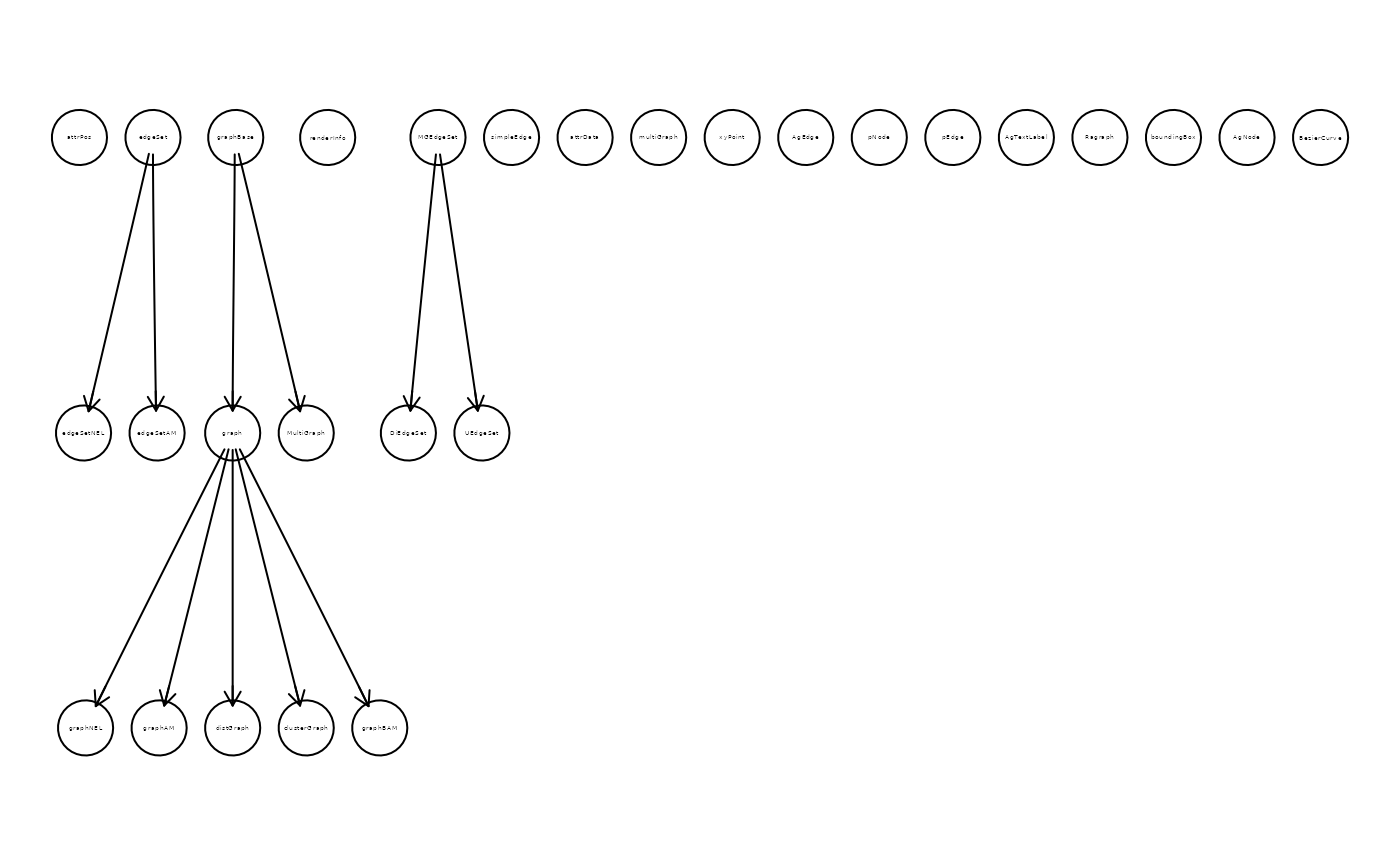
adjacencyOfClasses("graph")
ac1 <- adjacencyOfClasses("graph", FALSE, "adjacencyMatrix")
## note the use of t() below
gr_ac1 <- graph::graphAM(adjMat = t(ac1$AM), edgemode = "directed")
if(require("Rgraphviz", quietly = TRUE, warn.conflicts = FALSE))
plot(gr_ac1)
## more than one package
ac2 <- adjacencyOfClasses(c("graph", "Rgraphviz"), FALSE, "adjacencyMatrix")
gr_ac2 <- graph::graphAM(adjMat = t(ac2$AM), edgemode = "directed")
if(require("Rgraphviz", quietly = TRUE))
plot(gr_ac2)
})
#> > adjacencyOfClasses("graph")
#> $vertices
#> [1] "simpleEdge" "multiGraph" "graphAM" "edgeSetNEL" "attrPos"
#> [6] "MGEdgeSet" "edgeSet" "DiEdgeSet" "distGraph" "renderInfo"
#> [11] "attrData" "graphNEL" "UEdgeSet" "edgeSetAM" "clusterGraph"
#> [16] "graphBase" "graph" "MultiGraph" "graphBAM"
#>
#> $edges
#> [1] "graph -> graphAM" "graph -> distGraph"
#> [3] "graph -> graphNEL" "graph -> clusterGraph"
#> [5] "graph -> graphBAM" "graphBase -> graph"
#> [7] "graphBase -> MultiGraph" "edgeSet -> edgeSetNEL"
#> [9] "edgeSet -> edgeSetAM" "MGEdgeSet -> DiEdgeSet"
#> [11] "MGEdgeSet -> UEdgeSet"
#>
#> > ac1 <- adjacencyOfClasses("graph", FALSE, "adjacencyMatrix")
#> > gr_ac1 <- graph::graphAM(adjMat = t(ac1$AM), edgemode = "directed")
#> > if (require("Rgraphviz", quietly = TRUE, warn.conflicts = FALSE)) plot(gr_ac1)
#>
#> Attaching package: ‘generics’
#> The following objects are masked from ‘package:base’:
#>
#> as.difftime, as.factor, as.ordered, intersect, is.element, setdiff,
#> setequal, union
#>
#> Attaching package: ‘BiocGenerics’
#> The following objects are masked from ‘package:stats’:
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from ‘package:base’:
#>
#> Filter, Find, Map, Position, Reduce, anyDuplicated, aperm, append,
#> as.data.frame, basename, cbind, colnames, dirname, do.call,
#> duplicated, eval, evalq, get, grep, grepl, is.unsorted, lapply,
#> mapply, match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
#> rank, rbind, rownames, sapply, saveRDS, table, tapply, unique,
#> unsplit, which.max, which.min
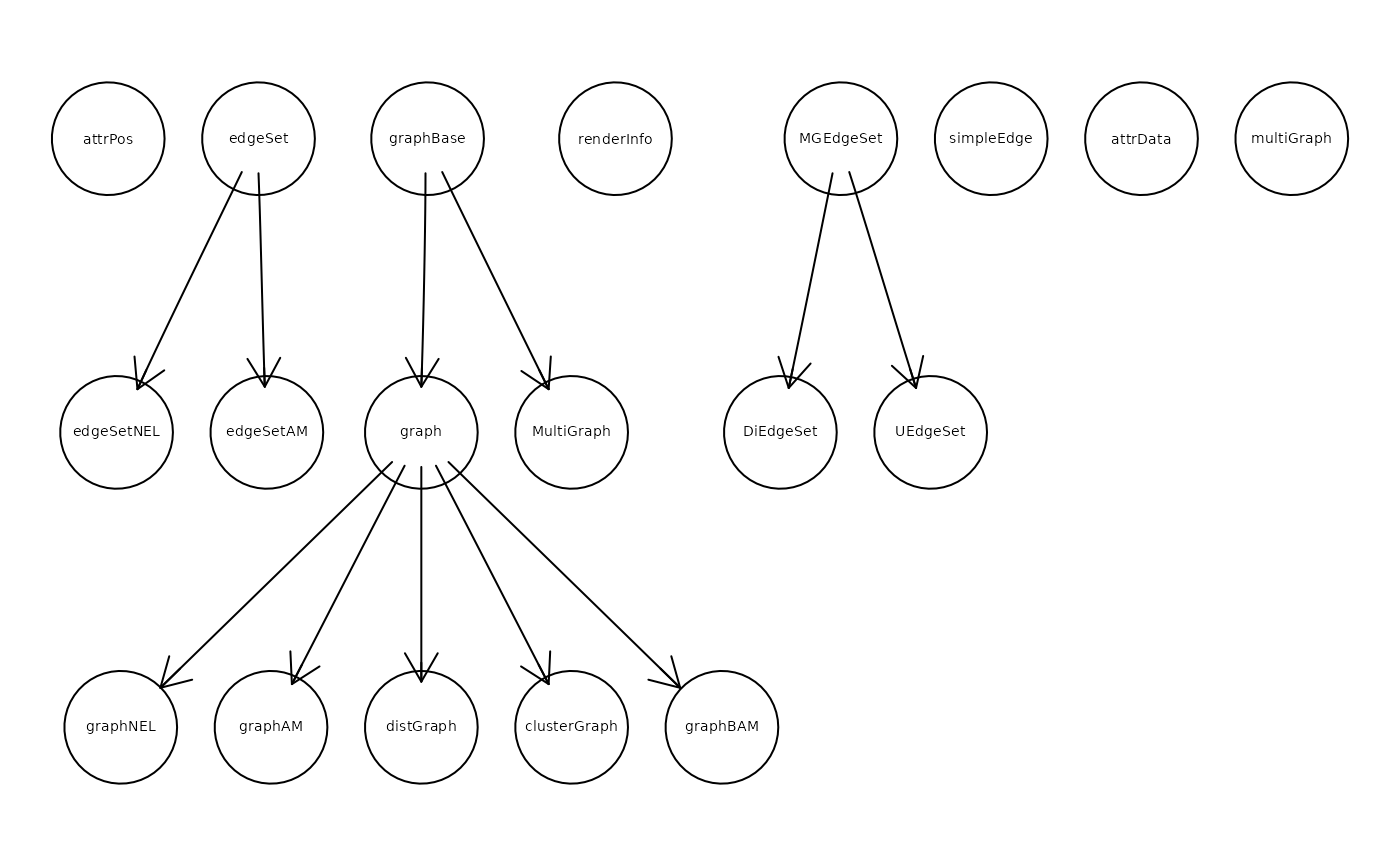 #> > ac2 <- adjacencyOfClasses(c("graph", "Rgraphviz"), FALSE, "adjacencyMatrix")
#> > gr_ac2 <- graph::graphAM(adjMat = t(ac2$AM), edgemode = "directed")
#> > if (require("Rgraphviz", quietly = TRUE)) plot(gr_ac2)
#> > ac2 <- adjacencyOfClasses(c("graph", "Rgraphviz"), FALSE, "adjacencyMatrix")
#> > gr_ac2 <- graph::graphAM(adjMat = t(ac2$AM), edgemode = "directed")
#> > if (require("Rgraphviz", quietly = TRUE)) plot(gr_ac2)