mixAR models with Gaussian noise components
MixARGaussian-class.RdClass "MixARGaussian" represents MixAR models with Gaussian
noise components.
Objects from the Class
Objects can be created by calls of the form new("MixARGaussian",
...), giving the elements of the model as named arguments, see the
examples below. All elements of the model, except arcoef, are
simple numeric vectors. From version 0.19-15 of package MixAR it is
possible to create objects using MixARGaussian(...). The two
forms are completely equivalent.
arcoef contains the AR coefficients, one numeric vector for
each mixture component. It can be given as a
"raggedCoef" object or as a list of numeric
vectors.
To input a model with seasonal AR coefficients, argument passed to arcoef can be passed as a raggedCoefS object, or as a list of three elements.
For the latter, seasonality s must be explicitly indicated.
AR coefficients can be given as list or matrix within the main list (one for main AR coefficients, named a, and one for seasonal AR coefficients, as). Each row of a input matrix/element of the list denotes one component of the mixture.
If not named, initialisation takes the first passed element to be a and the second to be as.
The AR order of the model is inferred from arcoef
argument. If argument order is given, it is checked for
consistency with arcoef. The shift slot defaults to a
vector of zeroes and the scale slot to a vector of ones.
The distribution of the noise components is standard Gaussian, N(0,1).
Slots
All slots except arcoef are numeric vectors of length
equal to the number of components in the model.
prob:probabilities of the mixture components
order:AR orders of the components
shift:the shift (intercept) terms of the AR components
scale:the standard deviations of the noise terms of the AR components
arcoef:The AR components, object of class
"raggedCoef"
Extends
Class "MixAR", directly.
Methods
- mix_cdf
signature(model = "MixARGaussian", x = "numeric", index = "numeric", xcond = "missing"): ...- mix_cdf
signature(model = "MixARGaussian", x = "numeric", index = "missing", xcond = "numeric"): ...- fit_mixAR
signature(x = "ANY", model = "MixARGaussian", init = "MixAR"): ...- get_edist
signature(model = "MixARGaussian"): ...- mix_cdf
signature(model = "MixARGaussian", x = "missing", index = "missing", xcond = "numeric"): ...- mix_pdf
signature(model = "MixARGaussian", x = "missing", index = "missing", xcond = "numeric"): ...- mix_pdf
signature(model = "MixARGaussian", x = "numeric", index = "missing", xcond = "numeric"): ...- mix_pdf
signature(model = "MixARGaussian", x = "numeric", index = "numeric", xcond = "missing"): ...- noise_dist
signature(model = "MixARGaussian"): ...- noise_rand
signature(model = "MixARGaussian"): ...
Examples
showClass("MixARGaussian")
#> Class "MixARGaussian" [package "mixAR"]
#>
#> Slots:
#>
#> Name: prob order shift scale arcoef
#> Class: numeric numeric numeric numeric raggedCoef
#>
#> Extends: "MixAR"
## load ibm data from BJ
## data(ibmclose, package = "fma")
## compute a predictive density, assuming exampleModels$WL_ibm model
## for the first date after the end of the data
pdf1 <- mix_pdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose))
#> Registered S3 method overwritten by 'quantmod':
#> method from
#> as.zoo.data.frame zoo
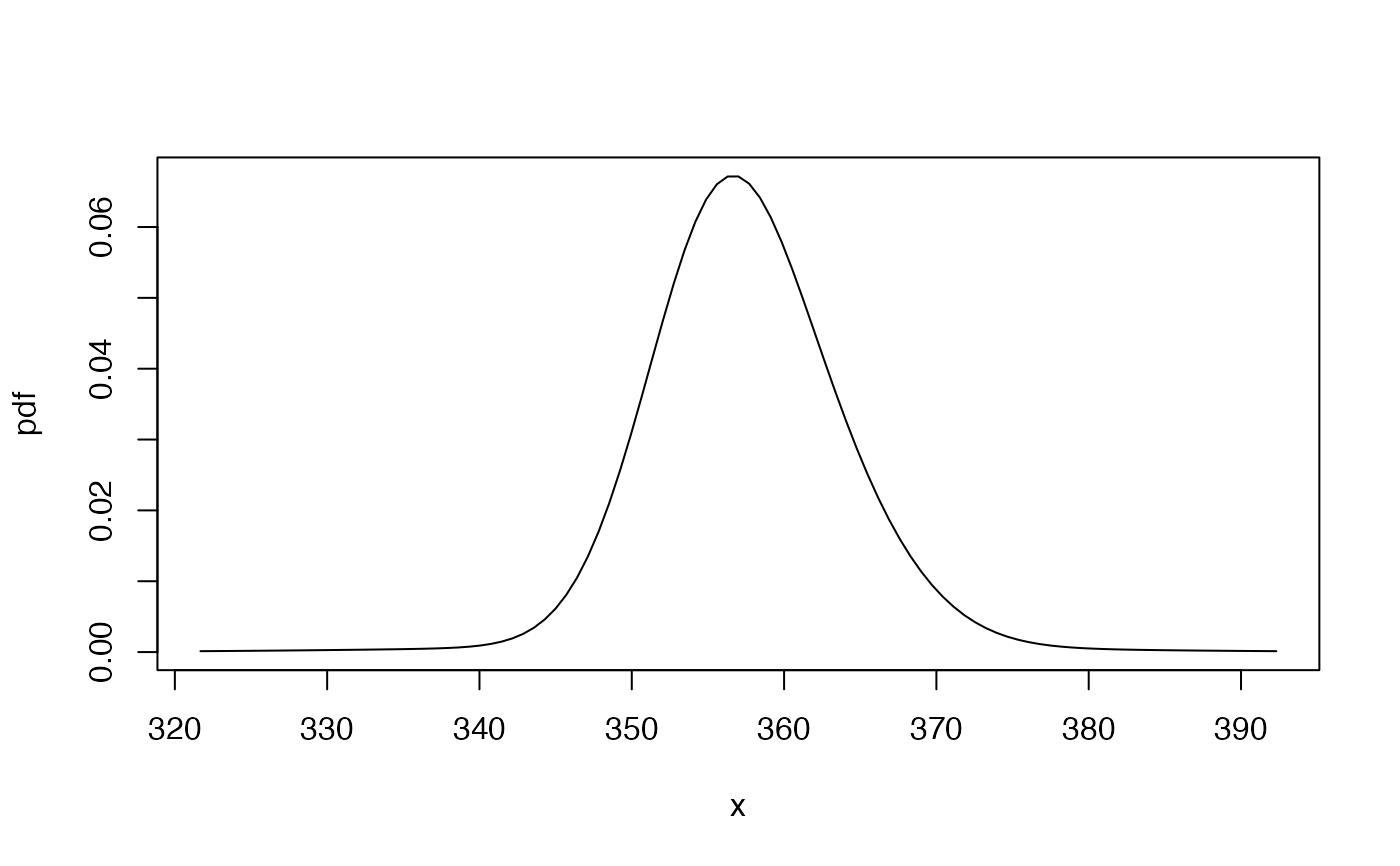
## plot the predictive density
## (cdf is used to determine limits on the x-axis)
cdf1 <- mix_cdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose))
gbutils::plotpdf(pdf1, cdf = cdf1, lq = 0.001, uq = 0.999)
 ## compute lower 5% quantile of cdf1
gbutils::cdf2quantile(0.05, cdf = cdf1)
#> [1] 347.7531
## compute lower 5% quantile of cdf1
gbutils::cdf2quantile(0.05, cdf = cdf1)
#> [1] 347.7531