Class "MixARgen"
MixARgen-class.RdA class for MixAR models with arbitrary noise distributions. "MixARgen"
inherits from "MixAR".
Objects from the Class
Objects can be created by calls of the form new("MixARgen",
dist, ...) or mixARgen(...). The two forms are completely
equivalent. The latter is available from version 0.19-15 of package
MixAR.
Slots
Most slots are inherited from class "MixAR".
prob:the mixing probabilities,
"numeric".order:the AR orders,
"numeric".shift:intercept terms,
"numeric".scale:scaling factor,
"numeric".arcoef:autoregressive coefficients, an object from class
"raggedCoef"containing one row for each mixture component.dist:Object of class
"list", representing the noise distributions. The list contains one element for each component of the MixAR model or a single element if the noise distribution is the same for all components.If the distributions do not contain parameters (e.g. Gaussian or \(t_4\)) it is sufficient to give the list of functions in the element
distof the list.If the distributions do contain parameters the recommended arrangement is to give a list with components
generatorandparam, such that a callgenerator(param)should produce the required list of distributions.This is not finalised but if changed, backward compatibility with existing objects will be maintained.
Extends
Class "MixAR", directly.
Methods
- get_edist
signature(model = "MixARgen"): ...- initialize
signature(.Object = "MixARgen"): ...- lik_params
signature(model = "MixARgen"): ...- mix_cdf
signature(model = "MixARgen", x = "missing", index = "missing", xcond = "numeric"): ...- mix_cdf
signature(model = "MixARgen", x = "numeric", index = "missing", xcond = "numeric"): ...- mix_cdf
signature(model = "MixARgen", x = "numeric", index = "numeric", xcond = "missing"): ...- mix_pdf
signature(model = "MixARgen", x = "missing", index = "missing", xcond = "numeric"): ...- mix_pdf
signature(model = "MixARgen", x = "numeric", index = "missing", xcond = "numeric"): ...- mix_pdf
signature(model = "MixARgen", x = "numeric", index = "numeric", xcond = "missing"): ...- noise_dist
signature(model = "MixARgen"): ...- noise_params
signature(model = "MixARgen"): ...- noise_rand
signature(model = "MixARgen"): ...
Examples
showClass("MixARgen")
#> Class "MixARgen" [package "mixAR"]
#>
#> Slots:
#>
#> Name: dist prob order shift scale arcoef
#> Class: list numeric numeric numeric numeric raggedCoef
#>
#> Extends: "MixAR"
exampleModels$WL_ibm_gen@dist
#> [[1]]
#> [[1]]$pdf
#> [1] "dnorm"
#>
#> [[1]]$cdf
#> [1] "pnorm"
#>
#> [[1]]$rand
#> [1] "rnorm"
#>
#> [[1]]$logpdf
#> function (x)
#> log(dnorm(x))
#> <bytecode: 0x55d83f259b68>
#> <environment: 0x55d83f255c10>
#>
#> [[1]]$Fscore
#> function (x)
#> -x
#> <bytecode: 0x55d83f25d308>
#> <environment: 0x55d83f255c10>
#>
#> [[1]]$xFscore
#> function (x)
#> -x^2
#> <bytecode: 0x55d83f25cd58>
#> <environment: 0x55d83f255c10>
#>
#> [[1]]$Parscore
#> function ()
#> 0
#> <bytecode: 0x55d83f25c738>
#> <environment: 0x55d83f255c10>
#>
#> [[1]]$get_param
#> function ()
#> numeric()
#> <bytecode: 0x55d83f25c230>
#> <environment: 0x55d83f255c10>
#>
#> [[1]]$set_param
#> function ()
#> stop("Standard normal has no parameters to set.")
#> <bytecode: 0x55d83f25bcf0>
#> <environment: 0x55d83f255c10>
#>
#> [[1]]$any_param
#> function ()
#> param_flag
#> <bytecode: 0x55d83f25b778>
#> <environment: 0x55d83f255c10>
#>
#> [[1]]$show
#> function ()
#> "Standard normal distribution"
#> <bytecode: 0x55d83f25f0a0>
#> <environment: 0x55d83f255c10>
#>
#>
noise_dist(exampleModels$WL_ibm_gen, "cdf")
#> [[1]]
#> [1] "pnorm"
#>
noise_dist(exampleModels$WL_ibm_gen, "pdf")
#> [[1]]
#> [1] "dnorm"
#>
noise_dist(exampleModels$WL_ibm_gen, "pdf", expand = TRUE)
#> [[1]]
#> [1] "dnorm"
#>
#> [[2]]
#> [1] "dnorm"
#>
#> [[3]]
#> [1] "dnorm"
#>
noise_dist(exampleModels$WL_ibm_gen, "cdf", expand = TRUE)
#> [[1]]
#> [1] "pnorm"
#>
#> [[2]]
#> [1] "pnorm"
#>
#> [[3]]
#> [1] "pnorm"
#>
## data(ibmclose, package = "fma") # for `ibmclose'
pdf1 <- mix_pdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose))
cdf1 <- mix_cdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose))
gbutils::plotpdf(pdf1, cdf = cdf1, lq = 0.001, uq = 0.999)
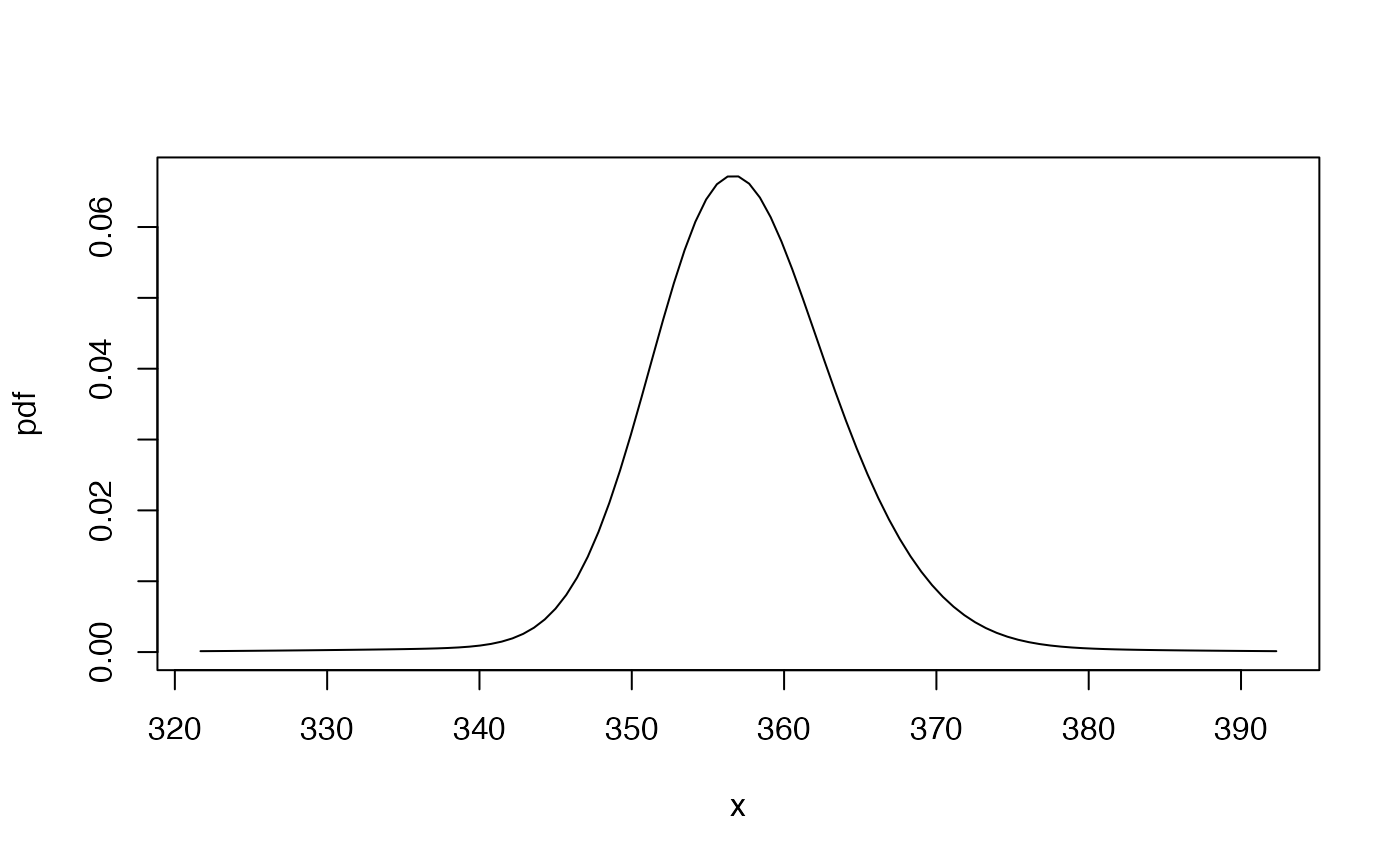 pdf1gen <- mix_pdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose))
cdf1gen <- mix_cdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose))
gbutils::plotpdf(pdf1gen, cdf = cdf1gen, lq = 0.001, uq = 0.999)
length(fma::ibmclose)
#> [1] 369
cdf1gena <- mix_cdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose)[-(369:369)])
pdf1gena <- mix_pdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose)[-(369:369)])
gbutils::plotpdf(pdf1gena, cdf = cdf1gena, lq = 0.001, uq = 0.999)
pdf1gen <- mix_pdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose))
cdf1gen <- mix_cdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose))
gbutils::plotpdf(pdf1gen, cdf = cdf1gen, lq = 0.001, uq = 0.999)
length(fma::ibmclose)
#> [1] 369
cdf1gena <- mix_cdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose)[-(369:369)])
pdf1gena <- mix_pdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose)[-(369:369)])
gbutils::plotpdf(pdf1gena, cdf = cdf1gena, lq = 0.001, uq = 0.999)
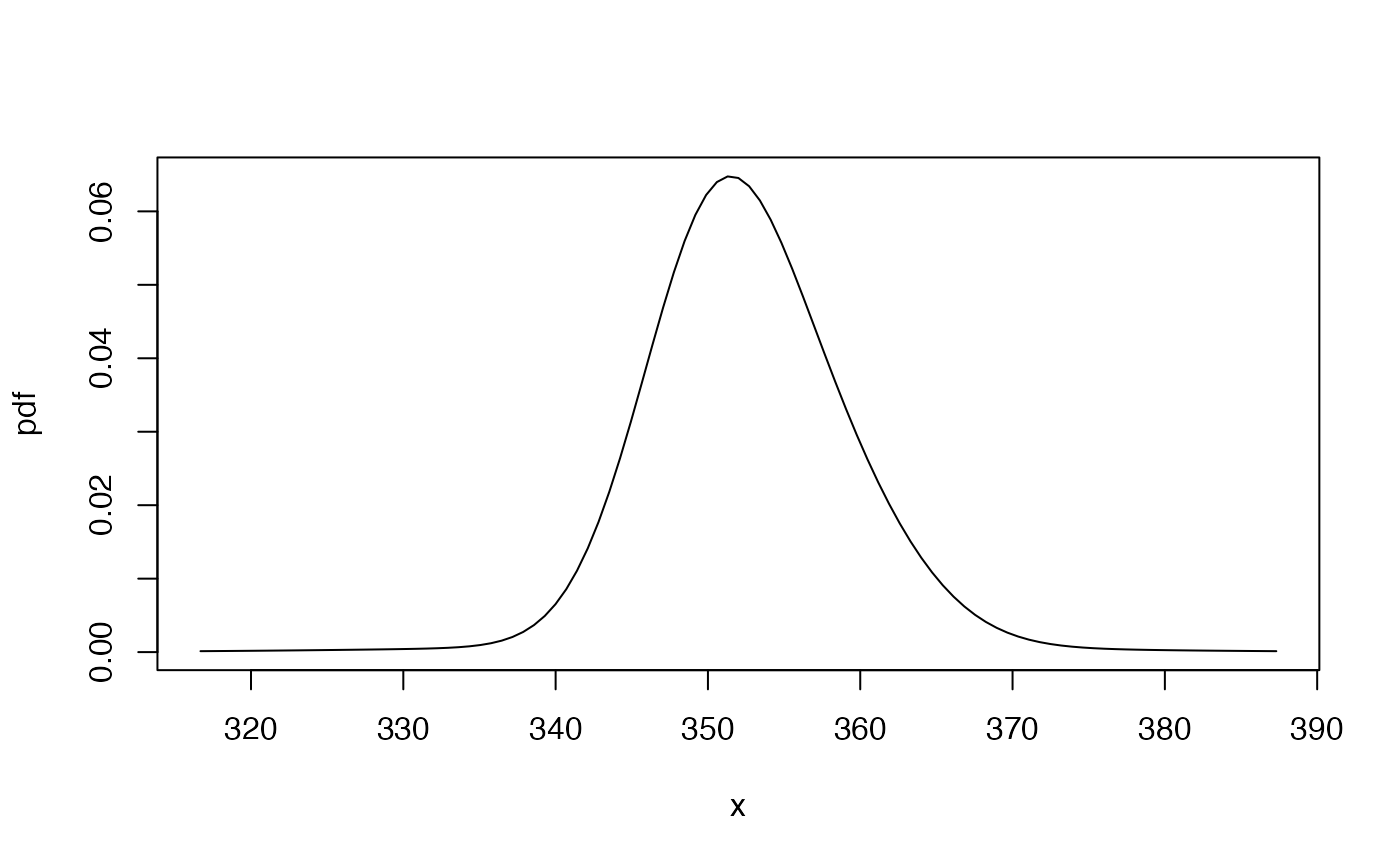 pdf1a <- mix_pdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose)[-(369:369)])
cdf1a <- mix_cdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose)[-(369:369)])
gbutils::plotpdf(pdf1a, cdf = cdf1a, lq = 0.001, uq = 0.999)
cdf1gena <- mix_cdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose)[-(369:369)])
cond_loglik(exampleModels$WL_ibm, as.numeric(fma::ibmclose))
#> [1] -1212.188
cond_loglik(exampleModels$WL_ibm_gen, as.numeric(fma::ibmclose))
#> [1] -1212.188
ts1gen <- mixAR_sim(exampleModels$WL_ibm_gen, n = 30, init = c(346, 352, 357), nskip = 0)
plot(ts1gen)
pdf1a <- mix_pdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose)[-(369:369)])
cdf1a <- mix_cdf(exampleModels$WL_ibm, xcond = as.numeric(fma::ibmclose)[-(369:369)])
gbutils::plotpdf(pdf1a, cdf = cdf1a, lq = 0.001, uq = 0.999)
cdf1gena <- mix_cdf(exampleModels$WL_ibm_gen, xcond = as.numeric(fma::ibmclose)[-(369:369)])
cond_loglik(exampleModels$WL_ibm, as.numeric(fma::ibmclose))
#> [1] -1212.188
cond_loglik(exampleModels$WL_ibm_gen, as.numeric(fma::ibmclose))
#> [1] -1212.188
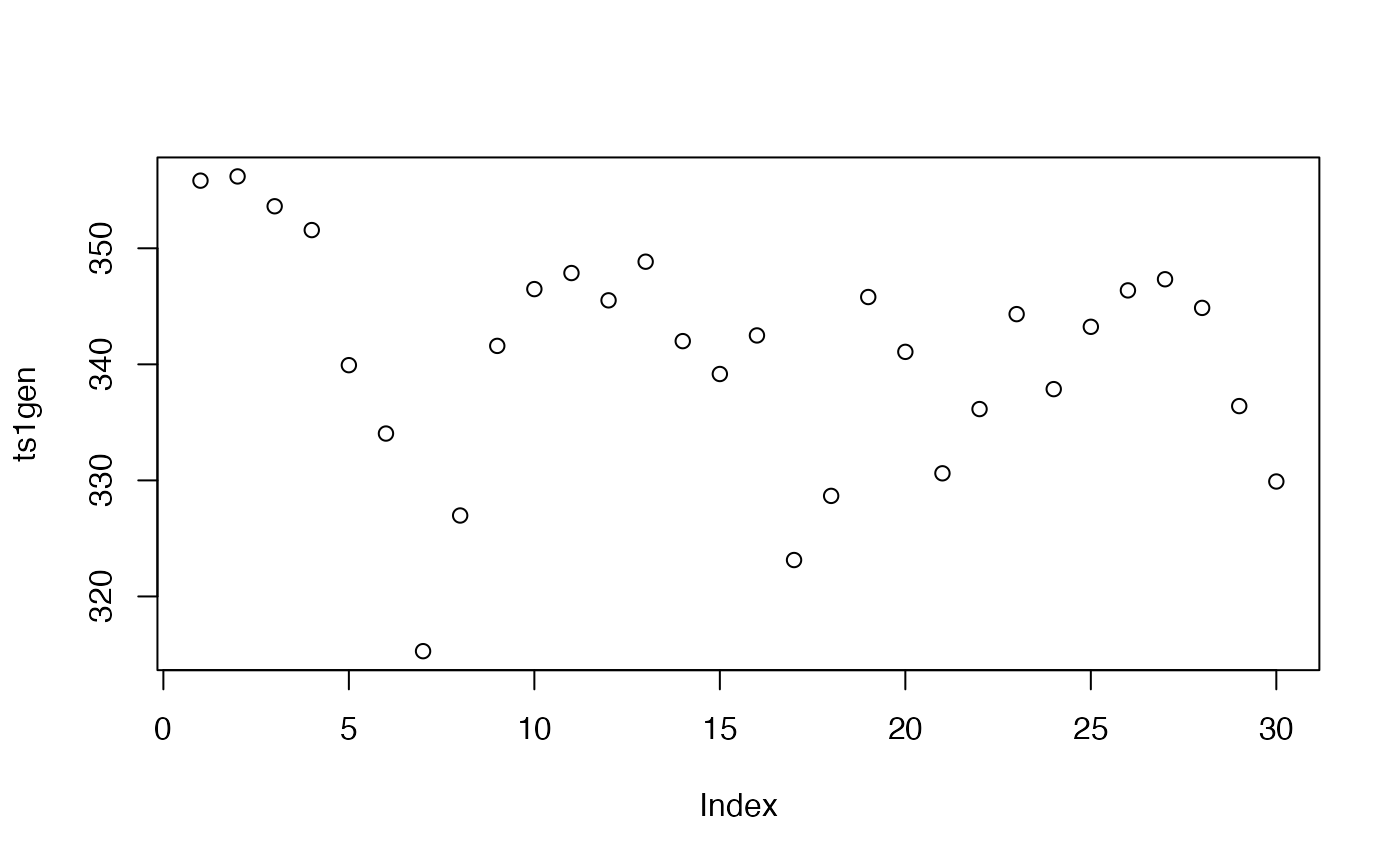
ts1gen <- mixAR_sim(exampleModels$WL_ibm_gen, n = 30, init = c(346, 352, 357), nskip = 0)
plot(ts1gen)
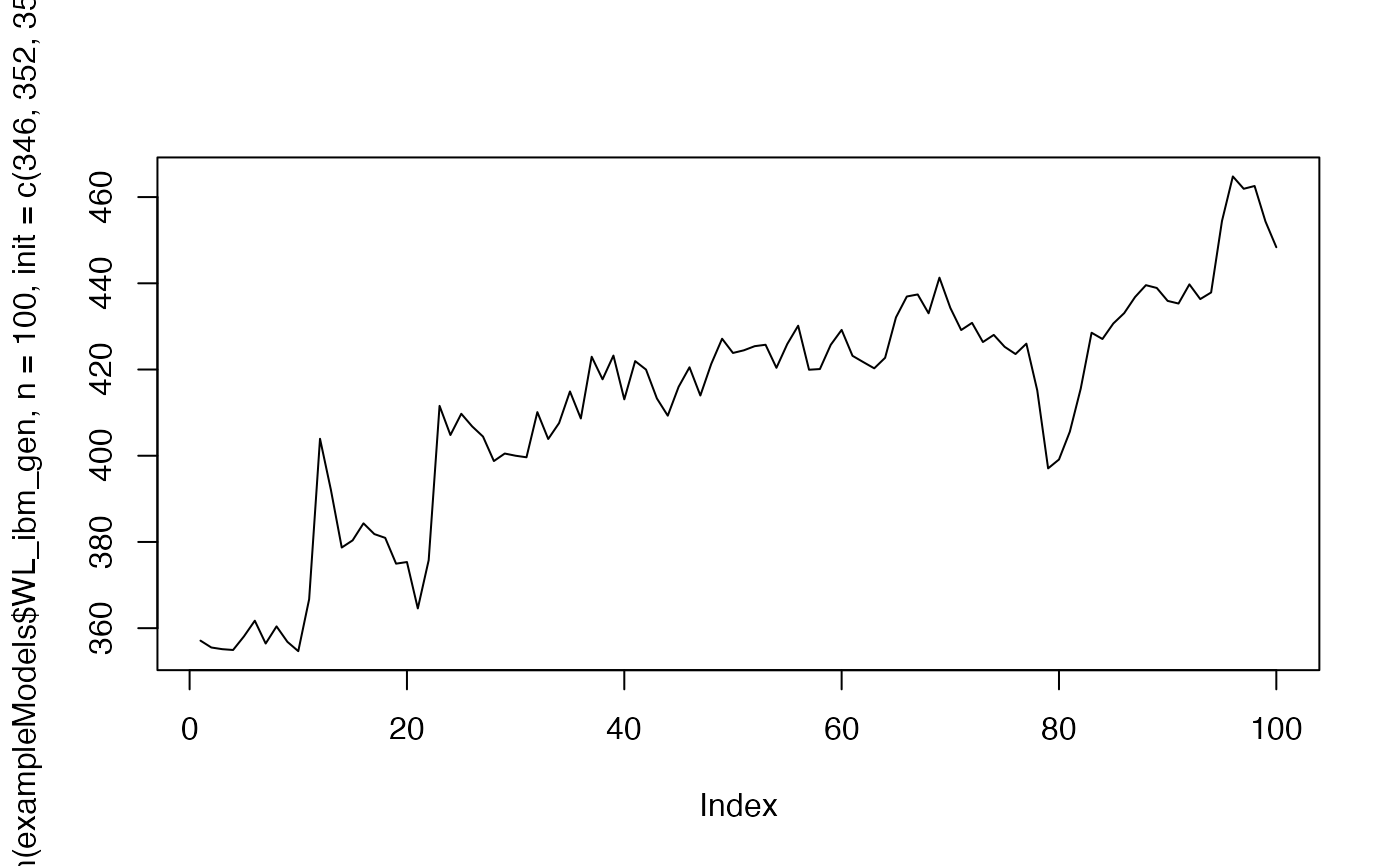
 plot(mixAR_sim(exampleModels$WL_ibm_gen, n = 100, init = c(346, 352, 357), nskip = 0),
type = "l")
plot(mixAR_sim(exampleModels$WL_ibm_gen, n = 100, init = c(346, 352, 357), nskip = 0),
type = "l")
 plot(diff(mixAR_sim(exampleModels$WL_ibm_gen, n = 100, init = c(346, 352, 357), nskip = 0)),
type = "l")
plot(diff(mixAR_sim(exampleModels$WL_ibm_gen, n = 100, init = c(346, 352, 357), nskip = 0)),
type = "l")
 noise_dist(exampleModels$WL_ibm_gen, "Fscore")
#> [[1]]
#> function (x)
#> -x
#> <bytecode: 0x55d83f25d308>
#> <environment: 0x55d83f255c10>
#>
prob <- exampleModels$WL_ibm@prob
scale <- exampleModels$WL_ibm@scale
arcoef <- exampleModels$WL_ibm@arcoef@a
mo_WLt3 <- new("MixARgen", prob = prob, scale = scale, arcoef = arcoef,
dist = list(fdist_stdt(3)))
mo_WLt30 <- new("MixARgen", prob = prob, scale = scale, arcoef = arcoef,
dist = list(fdist_stdt(30)))
noise_dist(exampleModels$WL_ibm_gen, "Fscore")
#> [[1]]
#> function (x)
#> -x
#> <bytecode: 0x55d83f25d308>
#> <environment: 0x55d83f255c10>
#>
prob <- exampleModels$WL_ibm@prob
scale <- exampleModels$WL_ibm@scale
arcoef <- exampleModels$WL_ibm@arcoef@a
mo_WLt3 <- new("MixARgen", prob = prob, scale = scale, arcoef = arcoef,
dist = list(fdist_stdt(3)))
mo_WLt30 <- new("MixARgen", prob = prob, scale = scale, arcoef = arcoef,
dist = list(fdist_stdt(30)))