Tests for weak white noise
acfGarchTest.RdCarry out tests for weak white noise under GARCH, GARCH-type, and stochastic volatility null hypotheses.
Arguments
- acr
autocorrelations.
- x
time series.
- nlags
how many lags to use.
- interval
If not NULL, compute also confidence intervals with the specified coverage probability.
- ...
additional arguments for the computation of the variance matrix under the null hypothesis, passed on to
nvarOfAcfKP.
Details
Unlike the autocorrelation IID test, the time series is needed here to estimate the covariance matrix of the autocorrelations under the null hypothesis.
acfGarchTest performs a test for uncorrelatedness of a time
series. The null hypothesis is that the time series is GARCH,
see Francq and Zakoian (2010)
.
acfWnTest performs a test for uncorrelatedness of a time
series under a weaker null hypothesis.
The null hypothesis is that the time series is GARCH-type or
from a stochasitc volatily model,
see Kokoszka and Politis (2011)
.
See the references for details and precise specification of the hypotheses.
The format of the return value is the same as for acfIidTest.
References
Francq C, Zakoian J (2010).
GARCH models: structure, statistical inference and financial applications.
John Wiley & Sons.
ISBN 978-0-470-68391-0.
Kokoszka PS, Politis DN (2011).
“Nonlinearity of ARCH and stochastic volatility models and Bartlett's formula.”
Probability and Mathematical Statistics, 31(1), 47--59.
See also
plot-methods for graphical representations of results
Examples
## see also the examples for \code{\link{whiteNoiseTest}}
set.seed(1234)
n <- 5000
x <- sarima:::rgarch1p1(n, alpha = 0.3, beta = 0.55, omega = 1, n.skip = 100)
x.acf <- autocorrelations(x)
x.pacf <- partialAutocorrelations(x)
acfGarchTest(x.acf, x = x, nlags = c(5,10,20))
#> $test
#> h Q pval
#> [1,] 5 2.867048 0.7204741
#> [2,] 10 9.064633 0.5259808
#> [3,] 20 24.420187 0.2245192
#>
#> $ci
#> int
#> [1,] -0.05923087 0.05923087
#> [2,] -0.05326386 0.05326386
#> [3,] -0.05066191 0.05066191
#> [4,] -0.04522221 0.04522221
#> [5,] -0.04507543 0.04507543
#> [6,] -0.04189048 0.04189048
#> [7,] -0.04210249 0.04210249
#> [8,] -0.03890700 0.03890700
#> [9,] -0.03517673 0.03517673
#> [10,] -0.03572033 0.03572033
#> [11,] -0.03155470 0.03155470
#> [12,] -0.03222239 0.03222239
#> [13,] -0.03105026 0.03105026
#> [14,] -0.03010171 0.03010171
#> [15,] -0.02875639 0.02875639
#> [16,] -0.02686352 0.02686352
#> [17,] -0.02775162 0.02775162
#> [18,] -0.02700132 0.02700132
#> [19,] -0.02673210 0.02673210
#> [20,] -0.02709457 0.02709457
#>
acfGarchTest(x.pacf, x = x, nlags = c(5,10,20))
#> $test
#> h Q pval
#> [1,] 5 2.884453 0.7177943
#> [2,] 10 9.183271 0.5148034
#> [3,] 20 27.271537 0.1277959
#>
#> $ci
#> int
#> [1,] -0.05923087 0.05923087
#> [2,] -0.05326386 0.05326386
#> [3,] -0.05066191 0.05066191
#> [4,] -0.04522221 0.04522221
#> [5,] -0.04507543 0.04507543
#> [6,] -0.04189048 0.04189048
#> [7,] -0.04210249 0.04210249
#> [8,] -0.03890700 0.03890700
#> [9,] -0.03517673 0.03517673
#> [10,] -0.03572033 0.03572033
#> [11,] -0.03155470 0.03155470
#> [12,] -0.03222239 0.03222239
#> [13,] -0.03105026 0.03105026
#> [14,] -0.03010171 0.03010171
#> [15,] -0.02875639 0.02875639
#> [16,] -0.02686352 0.02686352
#> [17,] -0.02775162 0.02775162
#> [18,] -0.02700132 0.02700132
#> [19,] -0.02673210 0.02673210
#> [20,] -0.02709457 0.02709457
#>
# do not compute CI's:
acfGarchTest(x.pacf, x = x, nlags = c(5,10,20), interval = NULL)
#> $test
#> h Q pval
#> [1,] 5 2.884453 0.7177943
#> [2,] 10 9.183271 0.5148034
#> [3,] 20 27.271537 0.1277959
#>
## plot methods call acfGarchTest() suitably if 'x' is given:
plot(x.acf, data = x)
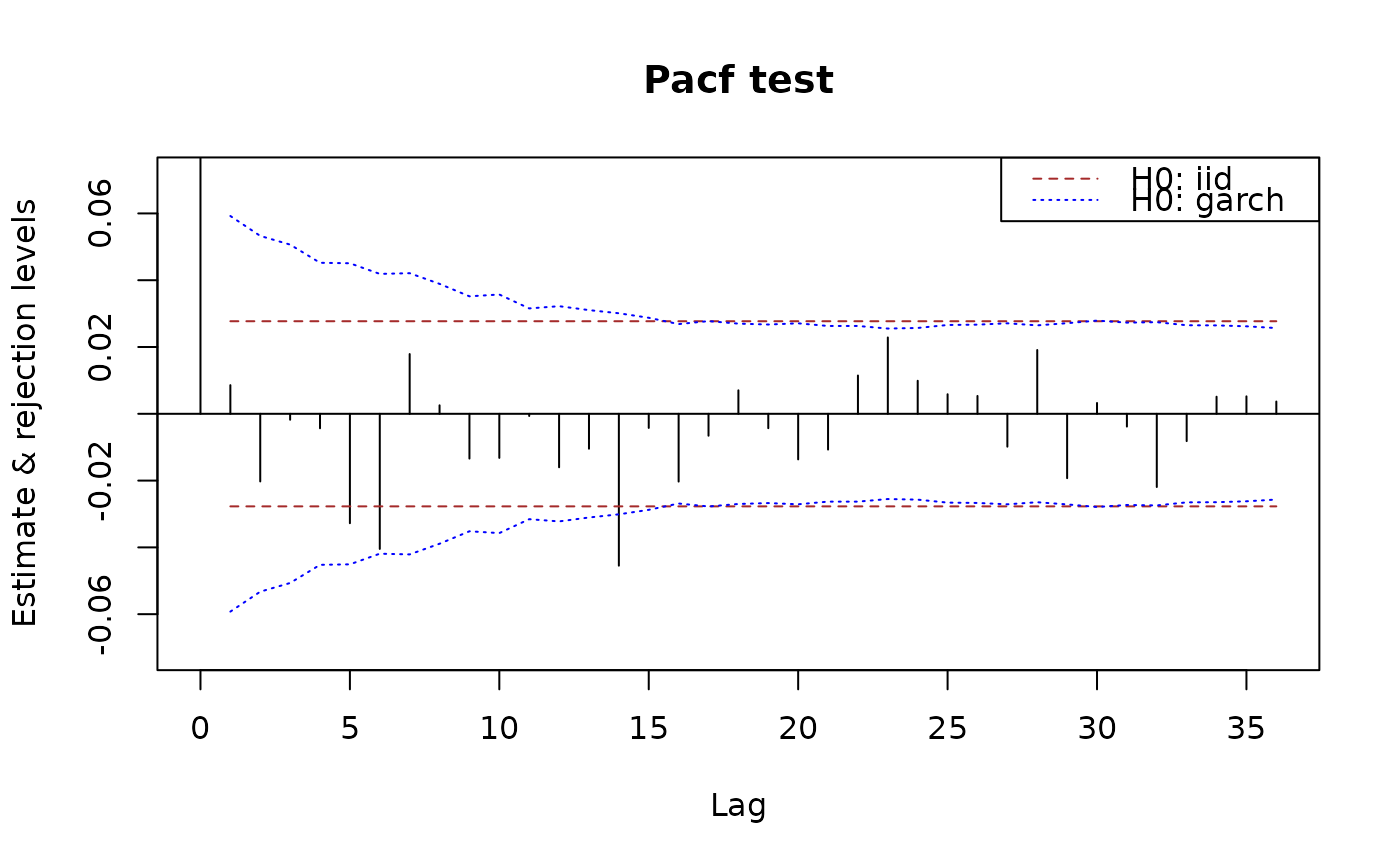
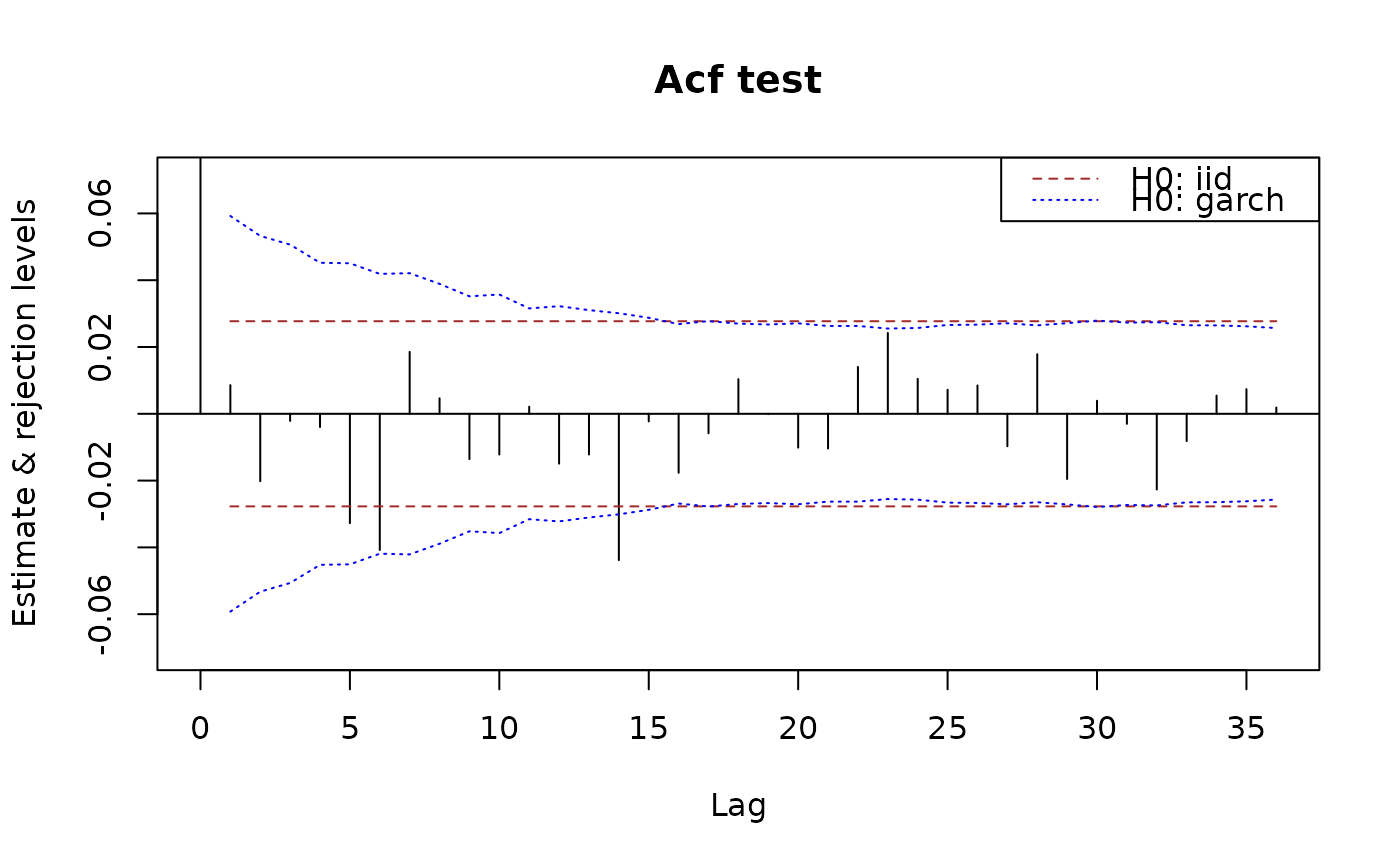 plot(x.pacf, data = x)
plot(x.pacf, data = x)
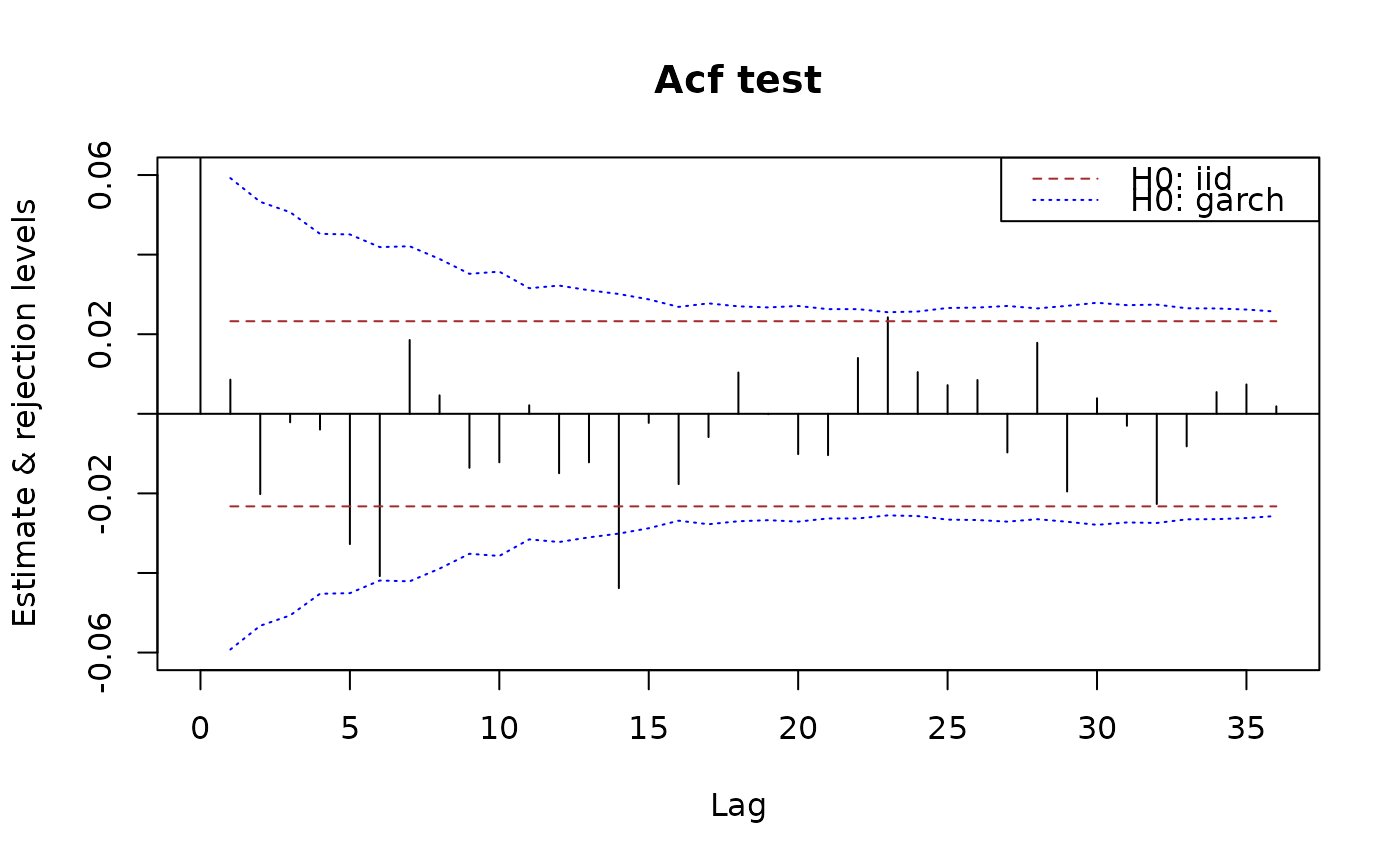
 ## use 90% limits:
plot(x.acf, data = x, interval = 0.90)
## use 90% limits:
plot(x.acf, data = x, interval = 0.90)
 acfWnTest(x.acf, x = x, nlags = c(5,10,20))
#> $test
#> h Q pval
#> [1,] 5 2.695352 0.7468293
#> [2,] 10 8.157398 0.6134657
#> [3,] 20 20.691112 0.4155097
#>
#> $ci
#> int
#> [1,] -0.05923087 0.05923087
#> [2,] -0.05326386 0.05326386
#> [3,] -0.05066191 0.05066191
#> [4,] -0.04522221 0.04522221
#> [5,] -0.04507543 0.04507543
#> [6,] -0.04189048 0.04189048
#> [7,] -0.04210249 0.04210249
#> [8,] -0.03890700 0.03890700
#> [9,] -0.03517673 0.03517673
#> [10,] -0.03572033 0.03572033
#> [11,] -0.03155470 0.03155470
#> [12,] -0.03222239 0.03222239
#> [13,] -0.03105026 0.03105026
#> [14,] -0.03010171 0.03010171
#> [15,] -0.02875639 0.02875639
#> [16,] -0.02686352 0.02686352
#> [17,] -0.02775162 0.02775162
#> [18,] -0.02700132 0.02700132
#> [19,] -0.02673210 0.02673210
#> [20,] -0.02709457 0.02709457
#>
nvarOfAcfKP(x, maxlag = 20)
#> [1] 4.5663595 3.6926586 3.3406961 2.6618118 2.6445613 2.2840442 2.3072224
#> [8] 1.9702857 1.6105891 1.6607521 1.2959906 1.3514172 1.2548862 1.1793864
#> [15] 1.0763228 0.9392902 1.0024221 0.9489510 0.9301220 0.9555168
whiteNoiseTest(x.acf, h0 = "arch-type", x = x, nlags = c(5,10,20))
#> $test
#> h Q pval
#> [1,] 5 2.695352 0.7468293
#> [2,] 10 8.157398 0.6134657
#> [3,] 20 20.691112 0.4155097
#>
#> $ci
#> int
#> [1,] -0.05923087 0.05923087
#> [2,] -0.05326386 0.05326386
#> [3,] -0.05066191 0.05066191
#> [4,] -0.04522221 0.04522221
#> [5,] -0.04507543 0.04507543
#> [6,] -0.04189048 0.04189048
#> [7,] -0.04210249 0.04210249
#> [8,] -0.03890700 0.03890700
#> [9,] -0.03517673 0.03517673
#> [10,] -0.03572033 0.03572033
#> [11,] -0.03155470 0.03155470
#> [12,] -0.03222239 0.03222239
#> [13,] -0.03105026 0.03105026
#> [14,] -0.03010171 0.03010171
#> [15,] -0.02875639 0.02875639
#> [16,] -0.02686352 0.02686352
#> [17,] -0.02775162 0.02775162
#> [18,] -0.02700132 0.02700132
#> [19,] -0.02673210 0.02673210
#> [20,] -0.02709457 0.02709457
#>
acfWnTest(x.acf, x = x, nlags = c(5,10,20))
#> $test
#> h Q pval
#> [1,] 5 2.695352 0.7468293
#> [2,] 10 8.157398 0.6134657
#> [3,] 20 20.691112 0.4155097
#>
#> $ci
#> int
#> [1,] -0.05923087 0.05923087
#> [2,] -0.05326386 0.05326386
#> [3,] -0.05066191 0.05066191
#> [4,] -0.04522221 0.04522221
#> [5,] -0.04507543 0.04507543
#> [6,] -0.04189048 0.04189048
#> [7,] -0.04210249 0.04210249
#> [8,] -0.03890700 0.03890700
#> [9,] -0.03517673 0.03517673
#> [10,] -0.03572033 0.03572033
#> [11,] -0.03155470 0.03155470
#> [12,] -0.03222239 0.03222239
#> [13,] -0.03105026 0.03105026
#> [14,] -0.03010171 0.03010171
#> [15,] -0.02875639 0.02875639
#> [16,] -0.02686352 0.02686352
#> [17,] -0.02775162 0.02775162
#> [18,] -0.02700132 0.02700132
#> [19,] -0.02673210 0.02673210
#> [20,] -0.02709457 0.02709457
#>
nvarOfAcfKP(x, maxlag = 20)
#> [1] 4.5663595 3.6926586 3.3406961 2.6618118 2.6445613 2.2840442 2.3072224
#> [8] 1.9702857 1.6105891 1.6607521 1.2959906 1.3514172 1.2548862 1.1793864
#> [15] 1.0763228 0.9392902 1.0024221 0.9489510 0.9301220 0.9555168
whiteNoiseTest(x.acf, h0 = "arch-type", x = x, nlags = c(5,10,20))
#> $test
#> h Q pval
#> [1,] 5 2.695352 0.7468293
#> [2,] 10 8.157398 0.6134657
#> [3,] 20 20.691112 0.4155097
#>
#> $ci
#> int
#> [1,] -0.05923087 0.05923087
#> [2,] -0.05326386 0.05326386
#> [3,] -0.05066191 0.05066191
#> [4,] -0.04522221 0.04522221
#> [5,] -0.04507543 0.04507543
#> [6,] -0.04189048 0.04189048
#> [7,] -0.04210249 0.04210249
#> [8,] -0.03890700 0.03890700
#> [9,] -0.03517673 0.03517673
#> [10,] -0.03572033 0.03572033
#> [11,] -0.03155470 0.03155470
#> [12,] -0.03222239 0.03222239
#> [13,] -0.03105026 0.03105026
#> [14,] -0.03010171 0.03010171
#> [15,] -0.02875639 0.02875639
#> [16,] -0.02686352 0.02686352
#> [17,] -0.02775162 0.02775162
#> [18,] -0.02700132 0.02700132
#> [19,] -0.02673210 0.02673210
#> [20,] -0.02709457 0.02709457
#>